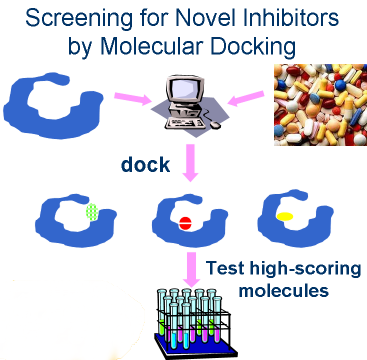
Virtual screening requires three things:
- a target (X-ray structure if available,
a comparative model otherwise)
- a database of small molecules (we use ZINC)
- a program to orient and score the molecules
in the protein binding site (we use DOCK 3.5.54, a
verison of UCSF DOCK).
There are good reasons why this approach might
not work (see
caveat lector)
such as problems in sampling relevant
poses, and problems with the scoring function. Despite these
problems, there are a growing number of success stories in
the literature using this technique. One reason it has been successful
is that failure is relatively cheap when using commercially
available libraries, such as ZINC. . If the first compound
purchased and tested does not work, it is a simple matter
to move on and test the next in a series of docking-prioritized
compounds, all of which are commercially available.
For more information of this technology in practice, see
success stories.
For more information in its possible problems, see
caveat lector.
For possible academic collaborations, and vendors of virtual screening
software, both outside of Blue Dolphin, see
other docking labs and software.